Research interests:
Bioinformatics / computational biology - medical bioinformatics with a primary focus on
(i) computational neuropathology, including cancer genetics/genomics of brain tumors as well as network analysis and the identification and prioritization of candidate genes for human hereditary disorders of the central nervous system; and
(ii) computational oncology in general, including the development and evolution of cancers in other organs or tissues.
Philosophy of biology - philosophical issues (loosly) associated with my primary research focus on computational biology; areas of interest include evolution (e.g. in the context of cancer), gene concepts and notions of essentialism, information and genetic programs in biology.
Past research focus:
Grid computing - distributed computing with a particular focus on grid accounting, resource usage prediction and economic approaches to job scheduling and workload balancing.
Education:
PhD in molecular medicine with focus on bioinformatics at the Molecular Biotechnology Center and the (former) Department of Genetics, Biology and Biochemistry of the University of Torino.
Graduate studies (Master's degree) in bioinformatics at University of Torino. Field of Study: Prioritization of candidate genes for human hereditary diseases.
Graduate studies (Laurea Specialistica) in physics at the University of Torino and the Italian National Institute of Nuclear Physics - Section of Torino (INFN Torino). Field of study: Grid computing in high energy physics; Economic Models for the Distributed Grid Accounting System (DGAS) of the European DataGrid (EDG).
Undergradute studies (Vordiplom and Laurea) in physics at the University of Heidelberg (Germany) and the University of Torino (Italy).
Professional experience:
Since February 2020: Associate Professor of Bioinformatics at the Department of Electronics, Information and Bioengineering (DEIB) of Politecnico di Milano, Italy.
Local technical coordinator of Politecnico di Milano's node of Elixir Italy (Italian Bioinformatics Infrastructure, IIB) (since January 2023).
Member of the Data-driven Genomic Computing (GeCo) project.
Assistant Professor (Juniorprofessor) of Bioinformatics (3/2015 to 2/2020) at the Dep. of Mathematics and Computer Sciene of Freie Universität Berlin and the Institute of Medical Genetics and Human Genetics at Charité-Universitätsmedizin Berlin, Germany.
Postdoctoral fellow (9/2013 to 2/2015) in the Division of Molecular Genetics (Headed by Prof. Peter Lichter) of the German Cancer Research Center (DKFZ) and the German Consortium for Translational Cancer Research (DKTK).
Postdoctoral fellow (3/2011 to 8/2013) in the former Division of Theoretical Bioinformatics (Headed by Prof. Roland Eils) of the German Cancer Research Center (DKFZ).
Researcher/programmer for INFN Torino (2/2005 to 8/2008) within the Middleware Reengineering & Integration task (JRA1) of the EGEE and EGEE-II projects (Enabling Grids for E-sciencE), within the Operations Service Activity (SA1) of the EGEE-III project, and within the JRA1 Accounting Activity of the OMII-Europe project (Open Middleware Infrastructure Institute for Europe). Coordination of the OMII-Europe project's JRA1 Accounting Activity task (4/2006 to 11/2006). [Previously: member of the European DataGrid (EDG) project (4/2002 to 3/2004) as an undergraduate student for INFN Torino and University of Torino.]
Tasks: Research and development as well as improvement of grid middleware for resource usage accounting, namely the Distributed Grid Accounting System (DGAS). For several years co-chair of the Resource Usage Service (RUS) working group and member of the Usage Record (UR) working group of the Open Grid Forum (OGF) to promote standardization, above all regarding issues related to accounting in grid environments.
Software developer at DCS Software e Servizi S.r.l. (6/2003 to 9/2004). Development of web and stand-alone applications for lawyers associations ("ordini degli avvocati"), e.g. for the management of and online access to sentences in digital form, in particular for the lawyers associations of Milan, Turin and Florence.
Student researcher/programmer (10/1997 to 7/1999 and 10/2000 to 3/2001) in the Division of Molecular Biophysics of the German Cancer Research Center (Deutsches Krebsforschungszentrum - DKFZ, Heidelberg). Responsibilities included the maintenance and improvement - in the C and Fortran programming languages - of programs for genetic sequence analysis and their adoption to HUSAR (Heidelberg Unix Sequence Analysis Resources).
About me in numbers
-
Erdős number: 4
-
[1] Peter J. Cameron coauthored with Paul Erdős [0]
[2] Christoph Buchheim coauthored with Peter J. Cameron
[3] Marcus Oswald coauthored with Christoph Buchheim
[4] Rosario M. Piro coauthored with Marcus Oswald
-
[1] Valentine Bargmann coauthored with Albert Einstein [0]
[2] Luciano Girardello coauthored with Valentine Bargmann
[3] Marco Billó coauthored with Luciano Girardello
[4] Paolo Provero coauthored with Marco Billó
[5] Rosario M. Piro coauthored with Paolo Provero
Publications
Highlights
2024
-
[Research article] Abdullahi MS, Suratanee A, Piro RM*, Plaimas K*. (*= senior authors)
Persistent Homology Identifies Pathways Associated with Hepatocellular Carcinoma from Peripheral Blood Samples.
Mathematics 12(5):725, 2024.
2023
-
[Research article] Reuss DE, Downing SM, Camacho CV, Wang Y-D, Piro RM, Herold-Mende C, Wang Z-Q, Hofmann TG, Sahm F, von Deimling A, McKinnon PJ, Frappart P-O.
Simultaneous Nbs1 and p53 inactivation in neural progenitors triggers High-Grade Gliomas (HGG).
Neuropathol Appl Neurobiol 49(4):e12915, 2023.
2021
-
[Research article] de Melo Costa VR, Pfeuffer J, Louloupi A, Ørom UAV, Piro RM.
SPLICE-q: a Python tool for genome-wide quantification of splicing efficiency.
BMC Bioinformatics 22:368, 2021.
-
[Research article] Epping L, Walther B, Piro RM, Knüver M-T, Huber C, Thürmer A, Flieger A, Fruth A, Janecko N, Wieler LH, Stingl K, Semmler T.
Genome-wide insights into population structure and host specificity of Campylobacter jejuni.
Scientific Reports 11(1):10358, 2021.
-
[Research article] Gulino A, Stamoulakatou E, Piro RM.
MutViz 2.0: visual analysis of somatic mutations and the impact of mutational signatures on selected genomic regions.
NAR Cancer 3(2):zcab012, 2021.
2020
-
[Book chapter / review article] Piro RM.
Sequencing technologies for epigenetics: from basics to applications.
Chapter 7 in: Kabelitz D, Bhat J (eds.) Epigenetics of the Immune System (Translational Epigenetics volume 16), pp. 161-183. Academic Press (Elsevier). ISBN: 978-0-1281-7964-2.
-
[Research article] Soni H, Bode J, Nguyen CDL, Puccio L, Neßling M, Piro RM, Bub J, Phillips E, Ahrends R, Eipper BA, Tews B, Goidts V.
PERK-mediated expression of peptidylglycine α-amidating monooxygenase supports angiogenesis in glioblastoma.
Oncogenesis 9(2):18, 2020.
-
[Research article] Stamoulakatou E, Pinoli P, Ceri S, Piro RM.
Impact of mutational signatures on microRNA and their response elements.
Pac Symp Biocomput 25:250-261, 2020.
2019
-
[Research article] Pinoli P, Stamoulakatou E, Ceri S, Piro RM.
Deleterious impact of mutational processes on transcription factor binding sites in human cancer.
Proceedings of the 19th IEEE International Conference on Bioinformatics and Bioengineering (BIBE), pp. 188-193, Athens, Greece, October 28-30, 2019.
-
[Research article] Krüger S, Piro RM.
decompTumor2Sig: identification of mutational signatures active in individual tumors.
BMC Bioinformatics 20(Suppl 4):152, 2019.
-
[Book chapter / review article] Piro RM, Marsico A.
Network-based methods and other approaches for predicting lncRNA functions and disease associations.
Methods in Molecular Biology 1912:301-321, 2019.
Chapter 12 in: Lai X, Gupta SK, Vera J (eds.), Computational Biology of Non-Coding RNA (MIMB, volume 1912), pp. 301-321. Humana Press/Springer, New York, NY, USA. ISBN: 978-1-4939-8981-2.
2018
-
[Research article] Gröbner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA, Johann PD, Balasubramanian GP, Segura-Wang M, Brabetz S, Bender S, Hutter B, Sturm D, Pfaff E, Hübschmann D, Zipprich G, Heinold M, Eils J, Lawerenz C, Erkek S, Lambo S, Waszak S, Blattmann C, Borkhardt A, Kuhlen M, Eggert A, Fulda S, Gessler M, Wegert J, Kappler R, Baumhoer D, Burdach S, Kirschner-Schwabe R, Kontny U, Kulozik AE, Lohmann D, Hettmer S, Eckert C, Bielack S, Nathrath M, Niemeyer C, Richter GH, Schulte J, Siebert R, Westermann F, Molenaar JJ, Vassal G, Witt H, Burkhardt B, Kratz CP, Witt O, van Tilburg CM, Kramm CM, Fleischhack G, Dirksen U, Rutkowski S, Frühwald M, von Hoff K, Wolf S, Klingebiel T, Koscielniak E, Landgraf P, Koster J, Resnick AC, Zhang J, Liu Y, Zhou X, Waanders AJ, Zwijnenburg DA, Raman P, Brors B, Weber UD, Northcott PA, Pajtler KW, Kool M, Piro RM, Korbel JO, Schlesner M, Eils R, Jones DTW, Lichter P, Chavez L, Zapatka M, Pfister SM; ICGC PedBrain-Seq Project; ICGC MMML-Seq Project.
The landscape of genomic alterations across childhood cancers.
Nature 555(7696):321-327, 2018.
2017
-
[Extended abstract] Krüger S, Piro RM.
Identification of mutational signatures active in individual tumors.
Proceedings of the NETTAB 2017 Workshop on Methods, Tools & Platforms for Personalized Medicine in the Big Data Era, Palermo, Italy, October 16-18, 2017.
PeerJ Preprints 5:e3257v1, 2017.
-
[Book] Robinson PN, Piro RM, Jäger M.
Computational Exome and Genome Analysis.
Chapman & Hall/CRC Mathematical and Computational Biology series. CRC Press (Taylor & Francis Group), Boca Raton, FL, USA. ISBN: 978-1-4987-7598-4.
-
[Research article] Schweizer Y, Meszaros Z, Jones DTW, Koelsche C, Boudalil M, Fiesel P, Schrimpf D, Piro RM, Brehmer S, von Deimling A, Kerl U, Seiz-Rosenhagen M, Capper D.
Molecular transition of an adult low-grade brain tumor to an atypical teratoid/rhabdoid tumor over a time-course of 14 years.
Journal of Neuropathology & Experimental Neurology 76(8):655-664, 2017.
2016
-
[Research article] Hutter S, Piro RM, Waszak SM, Kehrer-Sawatzki H, Friedrich RE, Lassaletta A, Witt O, Korbel JO, Lichter P, Schuhmann MU, Pfister SM, Tabori U, Mautner VF, Jones DTW.
No correlation between NF1 mutation position and risk of optic pathway glioma in 77 unrelated NF1 patients.
Human Genetics 135(5):469-475, 2016.
2014
-
[Research article] Hutter S, Piro RM, Reuss DE, Hovestadt V, Sahm F, Farschtschi S, Kehrer-Sawatzki H, Wolf S, Lichter P, von Deimling A, Schuhmann MU, Pfister SM, Jones DT, Mautner VF.
Whole exome sequencing reveals that the majority of schwannomatosis cases remain unexplained after excluding SMARCB1 and LZTR1 germline variants.
Acta Neuropathologica 128(3):449-452, 2014.
-
[Research article] Piro RM, Wiesberg S, Schramm G, Rebel N, Oswald M, Eils R, Reinelt G, König R..
Network topology-based detection of differential gene regulation and regulatory switches in cell metabolism and signaling.
BMC Systems Biology 8:56, 2014.
-
[Research article] Kool M, Jones DT, Jäger N, Northcott PA, Pugh TJ, Hovestadt V, Piro RM, Esparza LA, Markant SL, Remke M, Milde T, Bourdeaut F, Ryzhova M, Sturm D, Pfaff E, Stark S, Hutter S, Seker-Cin H, Johann P, Bender S, Schmidt C, Rausch T, Shih D, Reimand J, Sieber L, Wittmann A, Linke L, Witt H, Weber UD, Zapatka M, König R, Beroukhim R, Bergthold G, van Sluis P, Volckmann R, Koster J, Versteeg R, Schmidt S, Wolf S, Lawerenz C, Bartholomae CC, von Kalle C, Unterberg A, Herold-Mende C, Hofer S, Kulozik AE, von Deimling A, Scheurlen W, Felsberg J, Reifenberger G, Hasselblatt M, Crawford JR, Grant GA, Jabado N, Perry A, Cowdrey C, Croul S, Zadeh G, Korbel JO, Doz F, Delattre O, Bader GD, McCabe MG, Collins VP, Kieran MW, Cho YJ, Pomeroy SL, Witt O, Brors B, Taylor MD, Schüller U, Korshunov A, Eils R, Wechsler-Reya RJ, Lichter P, Pfister SM; ICGC PedBrain Tumor Project.
Genome sequencing of SHH medulloblastoma predicts genotype-related response to Smoothened inhibition.
Cancer Cell 25(3):393-405, 2014.
-
[Research article] Mack SC*, Witt H*, Piro RM, Gu L, Zuyderduyn S, Stütz AM, Wang X, Gallo M, Garzia L, Zayne K, Zhang X, Ramaswamy V, Jäger N, Jones DT, Sill M, Pugh TJ, Ryzhova M, Wani KM, Shih DJ, Head R, Remke M, Bailey SD, Zichner T, Faria CC, Barszczyk M, Stark S, Seker-Cin H, Hutter S, Johann P, Bender S, Hovestadt V, Tzaridis T, Dubuc AM, Northcott PA, Peacock J, Bertrand KC, Agnihotri S, Cavalli FM, Clarke I, Nethery-Brokx K, Creasy CL, Verma SK, Koster J, Wu X, Yao Y, Milde T, Sin-Chan P, Zuccaro J, Lau L, Pereira S, Castelo-Branco P, Hirst M, Marra MA, Roberts SS, Fults D, Massimi L, Cho YJ, Van Meter T, Grajkowska W, Lach B, Kulozik AE, von Deimling A, Witt O, Scherer SW, Fan X, Muraszko KM, Kool M, Pomeroy SL, Gupta N, Phillips J, Huang A, Tabori U, Hawkins C, Malkin D, Kongkham PN, Weiss WA, Jabado N, Rutka JT, Bouffet E, Korbel JO, Lupien M, Aldape KD, Bader GD, Eils R, Lichter P, Dirks PB, Pfister SM, Korshunov A, Taylor MD. (*= first authors)
Epigenomic alterations define lethal CIMP-positive ependymomas of infancy.
Nature 506(7489):445-450, 2014.
-
[Research article] Bageritz J, Puccio L, Piro RM, Hovestadt V, Phillips E, Pankert T, Lohr J, Herold-Mende C, Lichter P, Goidts V.
Stem cell characteristics in glioblastoma are maintained by the ecto-nucleotidase E-NPP1.
Cell Death & Differentiation 21(6):929-940, 2014.
2013
-
[Research article] Tönjes M, Barbus S, Park YJ, Wang W, Schlotter M, Lindroth AM, Pleier SV, Bai AH, Karra D, Piro RM, Felsberg J, Addington A, Lemke D, Weibrecht I, Hovestadt V, Rolli CG, Campos B, Turcan S, Sturm D, Witt H, Chan TA, Herold-Mende C, Kemkemer R, König R, Schmidt K, Hull WE, Pfister SM, Jugold M, Hutson SM, Plass C, Okun JG, Reifenberger G, Lichter P, Radlwimmer B.
BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1.
Nature Medicine 19(7):901-908, 2013.
-
[Research article] Schweizer L*, Koelsche C*, Sahm F*, Piro RM*, Capper D, Reuss DE, Pusch S, Habel A, Meyer J, Göck T, Jones DT, Mawrin C, Schittenhelm J, Becker A, Heim S, Simon M, Herold-Mende C, Mechtersheimer G, Paulus W, König R, Wiestler OD, Pfister SM, von Deimling A. (*= first authors)
Meningeal hemangiopericytoma and solitary fibrous tumors carry the NAB2-STAT6 fusion and can be diagnosed by nuclear expression of STAT6 protein.
Acta Neuropathologica 125(5):651-658, 2013.
-
[Research article] Reuss DE*, Piro RM*, Jones DTW*, Simon M, Ketter R, Kool M, Becker A, Sahm F, Pusch S, Meyer J, Hagenlocher C, Schweizer L, Capper D, Kickingereder P, Mucha J, Koelsche C, Jäger N, Santarius T, Tarpey PS, Stephens PJ, Andrew Futreal P, Wellenreuther R, Kraus J, Lenartz D, Herold-Mende C, Hartmann C, Mawrin C, Giese N, Eils R, Collins VP, K&omul;nig R, Wiestler OD, Pfister SM, von Deimling A. (*= first authors)
Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations.
Acta Neuropathologica 125(3):351-358, 2013.
-
[Research article] Piro RM, Molineris I, Di Cunto F, Eils R, König R.
Disease-gene discovery by integration of 3D gene expression and transcription factor binding affinities.
Bioinformatics 29(4):468-475, 2013.
2012
-
[Review article] Piro RM.
Network medicine: linking disorders.
Human Genetics 131(12):1811-1820, 2012.
-
[Review article] Piro RM, Di Cunto F.
Computational approaches to disease-gene prediction: rationale, classification and successes.
FEBS Journal 279(5):678-696, 2012.
2011
-
[Research article] Piro RM, Molineris I, Ala U, Di Cunto F.
Evaluation of candidate genes from orphan FEB and GEFS+ loci by analysis of human brain gene expression atlases.
PLoS ONE 6(8):e23149, 2011.
-
[Research article] Piro RM*, Ala U*, Molineris I*, Grassi E, Bracco C, Perego GP, Provero P, Di Cunto F. (*= first authors)
An atlas of tissue-specific conserved coexpression for functional annotation and disease gene prediction.
European Journal of Human Genetics 19(11):1173-1180, 2011.
-
[Research article; philosophy] Piro RM.
Are all genes regulatory genes?
Biology & Philosophy 26(4):595-602, 2011. [Final draft: pdf]
-
[PhD thesis] Piro RM.
Computational analysis of spatially mapped gene expression and conserved co-expression for disease gene prediction
PhD in Molecular Medicine, University of Torino, January 2011.
2010
-
[Research article] Piro RM, Molineris I, Ala U, Provero P, Di Cunto F.
Candidate gene prioritization based on spatially mapped gene expression: an application to XLMR.
Bioinformatics 26(18):i618-624, 2010. [Presented at the European Conference on Computational Biology 2010 (ECCB10), September 26-29, Ghent, Belgium]
-
[Research article] Fagoonee S, Hobbs RM, De Chiara L, Cantarella D, Piro RM, Tolosano E, Medico E, Provero P, Pandolfi PP, Silengo L, Altruda F.
Generation of functional hepatocytes from mouse germline cell-derived pluripotent stem cells in vitro.
Stem Cells and Development 19(8):1183-1194, 2010.
-
[Book review; philosophy] Piro RM.
Né Kupiec né Sonigo
LN LibriNuovi 52(10.1):12-22, 2010. [Italian; pdf; out of print, see for example Book Depository]
-
[Book chapter / review article] Guarise A, Piro RM.
Accounting as a requirement for market-oriented Grid computing.
Chapter 9 in: Buyya, R. and Bubendorfer, K. (eds.), Market-Oriented Grid and Utility Computing (Wiley Series on Parallel and Distributed Computing), pp. 187-211. Wiley Press, Hoboken, New Jersey, USA. ISBN: 978-0-4702-8768-2.
2009
-
[Research article] Piro RM, Guarise A, Patania G, Werbrouck A.
Using historical accounting information to predict the resource usage of Grid jobs.
Future Generation Computer Systems 25(5):499-510, 2009.
-
[Book chapter / review article] Piro RM.
Resource usage accounting in Grid computing.
Chapter 18 in: Udoh, E. and Wang, F. (eds.), Handbook of Research on Grid Technologies and Utility Computing: Concepts for Managing Large-Scale Application (Encyclopedia of Grid Computing Technologies and Applications), pp. 183-193. Information Science Reference (IGI Global), Hershey, PA, USA. ISBN: 978-1-6056-6184-1.
2008
-
[Research article] Miozzi L, Piro RM, Rosa F, Ala U, Silengo L, Di Cunto F, Provero P.
Functional annotation and identification of candidate disease genes by computational analysis of normal tissue gene expression data.
PLoS ONE 3(6):e2439, 2008.
-
[Research article] Ala U*, Piro RM*, Grassi E, Damasco C, Silengo L, Oti M, Provero P, Di Cunto F. (*= first authors)
Prediction of human disease genes by human-mouse conserved coexpression analysis.
PLoS Computational Biology 4(3):e1000043, 2008.
-
[Research article] Andreetto P, Andreozzi S, Avellino G, Beco S, Cavallini A, Cecchi M, Ciaschini V, Dorise A, Giacomini F, Gianelle A, Grandinetti U, Guarise A, Krop A, Lops R, Maraschini A, Martelli V, Marzolla M, Mezzadri M, Molinari E, Monforte S, Pacini F, Pappalardo M, Parrini A, Patania G, Petronzio L, Piro R, Porciani M, Prelz F, Rebatto D, Ronchieri E, Sgaravatto M, Venturi V, Zangrando L.
The gLite Workload Management System.
Journal of Physics: Conference Series 119(6):062007, 2008. [Presented at: Computing in High Energy and Nuclear Physics (CHEP'07), Victoria, Canada, September 2-7, 2007.]
2007
-
[Research article] Piro RM*, Pace M*, Ghiselli A, Guarise A, Luppi E, Patania G, Tomassetti L, Werbrouck A. (*= first authors)
Tracing resource usage over heterogeneous Grid platforms: a prototype RUS interface for DGAS.
Proceedings of the 3rd International Conference on e-Science and Grid Computing (eScience 2007), pp. 93-100, Bangalore, India, December 10-13, 2007.
-
[Technical report] Riedel M, Netzer G, Piro RM
OGSA – Resource Usage Service DRAFT Charter.
Version 2007-01-26. Resource Usage Service Working Group (RUS-WG), Open Grid Forum (OGF), January 2007. [pdf]
2006
-
[Technical report] Piro RM, Canal P, Gordon J, Kant D, Khan A, Chen X.
Aggregate Usage Representation Recommendation.
Version 1.0. Usage Record Working Group (UR-WG), Open Grid Forum (OGF), December 2006. [pdf]
-
[Technical report] Piro RM.
Evaluation and Design Plan of a RUS Interface for DGAS.
Technical report. DRAFT (Version 0.2) - September 22, 2006. [pdf]
-
[Research article] Piro RM, Guarise A, Werbrouck A.
Price-sensitive resource brokering with the Hybrid Pricing Model and widely overlapping price domains.
Concurrency and Computation: Practice and Experience 18(8):837-850, 2006. [preprint: pdf]
-
[Master's thesis] Piro RM.
A method for prioritizing candidate genes for human hereditary diseases based on co-expression
Master's degree in Bioinformatics, University of Torino, April 2006.
-
[Research paper] Ciaschini V, Ferraro A, Ghiselli A, Rubini G, Guarise A, Patania G, Piro RM, Caltroni A.
An integrated framework for VO-oriented authorization, policy-based management and accounting.
Computing in High Energy and Nuclear Physics (CHEP'06), T.I.F.R. Mumbai, India, February 13-17, 2006.
-
[Research paper] Avellino G, Beco S, Cavallini A, Maraschini A, Pacini F, Parrini A, Scarcella C, Sottilaro M, Terracina A, Dvořák F, Kouřil D, Křenek A, Matyska L, Mulač M, Pospíšil J, Ruda M, Salvet Z, Sitera J, Škrabal J, Voců M, Monforte S, Pappalardo M, Andreozzi S, Cecchi M, Ciaschini V, Ferrari T, Giacomini F, Lops R, Ronchieri E, Venturi V, Fiorentino G, Martellli V, Mezzadri M, Molinari E, Prelz F, Rebatto D, Andreetto P, Borgia AS, Dorigo A, Giannelle A, Marzolla M, Mordacchini M, Sgaravatto M, Zangrando L, Guarise A, Patania G, Piro R, Werbrouck A.
Flexible job submission using Web Services: the gLite WMProxy experience.
Computing in High Energy and Nuclear Physics (CHEP'06), T.I.F.R. Mumbai, India, February 13-17, 2006.
-
[Research article] Andreetto P, Borgia SA, Dorigo A, Gianelle A, Marzolla M, Mordacchini M, Sgaravatto M, Zangrando L, Dvořák F, Kouřil D, Křenek A, Matyska L, Mulač M, Pospíšil J, Ruda M, Salvet Z, Sitera J, Škrabal J, Voců M, Avellino G, Beco S, Cavallini A, Maraschini A, Pacini F, Parrini A, Scarcella C, Sottilaro M, Terracina A, Monforte S, Pappalardo M, Andreozzi S, Cecchi M, Ciaschini V, Ferrari T, Giacomini F, Lops R, Ronchieri E, Venturi V, Fiorentino G, Martelli V, Mezzadri M, Molinari E, Prelz F, Rebatto D, Guarise A, Patania G, Piro R, Werbrouck A.
CREAM: a simple, Grid-accessible, job management system for local computational resources.
Computing in High Energy and Nuclear Physics (CHEP'06), T.I.F.R. Mumbai, India, February 13-17, 2006.
2004
-
[Research article] Avellino G, Beco S, Cantalupo B, Maraschini A, Pacini F, Sottilaro M, Terracina A, Colling D, Giacomini F, Ronchieri E, Gianelle A, Mazzucato M, Peluso R, Sgaravatto M, Guarise A, Piro R, Werbrouck A, Kouřil D, Křenek A, Matyska L, Mulač M, Pospíšil J, Ruda M, Salvet Z, Sitera J, Škrabal J, Voců M, Mezzadri M, Prelz F, Monforte S, Pappalardo M.
The DataGrid Workload Management System: challenges and results.
Journal of Grid Computing 2:353-367, 2004.
-
[Research article] Gianelle A, Peluso R, Sgaravatto M, Giacomini F, Ronchieri R, Avellino G, Cantalupo B, Beco S, Maraschini A, Pacini F, Guarise A, Piro R, Werbrouck A, Kouřil D, Křenek A, Kabela Z, Matyska L, Mulač M, Pospíšil J, Ruda M, Salvet Z, Sitera J, Voců M, Mezzadri M, Prelz F, Monforte S, Pappalardo M, D.Colling.
Supporting the development process of the DataGrid Workload Management System software with GNU autotools, CVS and RPM.
Computing in High Energy and Nuclear Physics (CHEP'04), Interlaken, Switzerland, September 27-October 1, 2004.
-
[Research article] Piro RM, Guarise A, Werbrouck A.
Simulation of price-sensitive resource brokering and the Hybrid Pricing Model with DGAS-Sim.
Proceedings of the 13th IEEE International Workshops on Enabling Technologies: Infrastructures for Collaborative Enterprises (WETICE-2004) [track on Emerging Technologies for Next generation GRID (ETNGRID 2004)], Modena, Italy, June 14-16, 2004.
-
[Master's thesis] Piro RM.
Simulation of economy-based load balancing in computational Grids for large-scale scientific applications
Master's degree (Laurea Specialistica) in Physics, University of Torino, April 2004.
2003
-
[Research article] Piro RM, Guarise A, Werbrouck A.
An economy-based accounting infrastructure for the DataGrid.
Proceedings of the 4th International Workshop on Grid Computing (GRID2003), Phoenix, Arizona (USA), November 17, 2003.
-
[Popular science article] Piro RM.
Un sistema di accounting economico per le grid computazionali / European DataGrid
NetworkNews Italia, numero 137, 5 dicembre 2003. [Italian; draft: pdf; final: pdf]
2002
-
[Bachelor thesis] Piro RM.
Development of a hardware and software-based test system for the power supply units of the CMS Silicon Strip Tracker
Bachelor (Laurea) in Physics, University of Torino, January 2002.
Downloads
CGPACE:
-
Piro RM, et al. Candidate gene prioritization based on spatially mapped gene expression: an application to XLMR. Bioinformatics 26(18):i618-624, 2010.
-
Piro RM, et al. Evaluation of candidate genes from orphan FEB and GEFS+ loci by analysis of human brain gene expression atlases. PLoS ONE 6(8):e23149, 2011.
-
Piro RM, et al. Disease-gene discovery by integration of 3D gene expression and transcription factor binding affinities. Bioinformatics 29(4):468-475, 2013.
CGPACE (Candidate Gene Prioritization by Analysis of Co-Expression) is the latest version of the software I developed and continusously improved for the following papers:
Note: despite its name, CGPACE can be used for data other than gene expression profiles (e.g., for total binding affinities/TBAs as in paper 3).
Citing CGPACE: Please cite paper 1 when using CGPACE with a Pearson correlation coefficient and paper 3 when using it with the Relative Intensity Overlap (RIO) and when using TBAs and/or a noisy-OR gate for data integration.
See the publications for descriptions of the algorithm, the RIO, the noisy-OR and the use of TBAs; contact me in case of further questions.
CGPACE is released under the terms of the GNU General Public License (Version 2 or later).
Stable release:
CGPACE-5-2012-12-03.tgz [source code, 57kB]; date: December, 2012.
decompTumor2Sig:
The decompTumor2Sig R package uses quadratic programming to decompose the mutation catalog from an individual tumor sample (or multiple individual tumor samples) into a set of given mutational signatures (either the Alexandrov model or the Shiraishi model), computing weights that reflect the contributions of the signatures to the mutation load of the tumor.
Citing decompTumor2Sig: Krüger S, Piro RM. decompTumor2Sig: identification of mutational signatures active in individual tumors. BMC Bioinformatics 20(Suppl 4):152, 2019.
DGAS-Sim:
DGAS-Sim(ulator) is a simple simulation tool of the architecture of the EDG Workload Management System (only the components involved in the scheduling process), written in Java, to study the impact of different resource pricing schemes on workload balancing.
DGAS-Sim is released under the terms of the EU DataGrid Software License.
Stable release:
edg-dgas-sim-0.1.0.tgz [source code, 50.2kB]; date: April, 2004.
Development release:
edg-dgas-sim-0.0.3dev.tgz [source code, 33.3kB]; date: September 24, 2003.
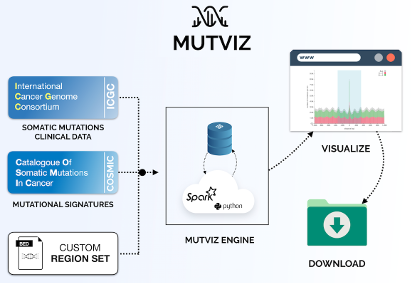
MutViz 2.0:
MutViz is a web tool which helps to identify enrichments of somatic mutations (single nucelotide variants) falling on predefined or user-specified sets of small genomic regions, such as transcription factor binding sites, promoters and others. MutViz is developed within the Data-driven Genomic Computing (GeCo) project.
Citing MutViz 2.0: Gulino A, Stamoulakatou E, Piro RM. MutViz 2.0: visual analysis of somatic mutations and the impact of mutational signatures on selected genomic regions. NAR Cancer 3(2):zcab012, 2021.
nnet:
nnet is a C++ library for artificial neural networks. Feed-forward networks with back-propagation are supported. Other network architectures may be derived from generic classes for neurons, synapses (connections) and networks.
nnet is released under the terms of the GNU General Public License (Version 2 or later).
It can also be downloaded from http://sourceforge.net/projects/nnet/.
Stable release:
nnet-0.1.0.tgz [source code, 43.5kB]; date: April 2, 2003.
PathWave:
-
Schramm G, et al. PathWave: Discovering patterns of differentially regulated enzymes in metabolic pathways. Bioinformatics 26(9):1225-1231, 2010.
- Piro RM, et al. Network topology-based detection of differential gene regulation and regulatory switches in cell metabolism and signaling. BMC Systems Biology 8:56, 2014.
PathWave enables the identification of disease-specific regulation patterns by combining gene expression data and network topology. By analyzing regulatory patterns at multiple scales, PAthWave is capabale to detect both global deregulation of pathways and localized regulatory switches that may still have a singificant impact on metabolic fluxes. PathWave is available here, or from our collaboration partners at Hans Knöll Institute (HKI), Jena:
Citing PathWave: Please cite Piro et al. (2014) for PathWave version 2.1, Schramm et al. (2010) for PathWave version 1.0, and both papers (Schramm et al. 2010; Piro et al. 2014) for PathWave in general (see PathWave→Publications for more details).
SPLICE-q:
SPLICE-q is a Python tool for genome-wide SPLICing Efficiency quantification from RNA-seq data. I takes a BAM file of mapped, strand-specific RNA-seq reads and a GTF gene annotation file and computes one of two available measurements of splicing efficiency for individual introns.
Citing SPLICE-q: de Melo Costa VR, Pfeuffer J, Louloupi A, Ørom UAV, Piro RM. SPLICE-q: a Python tool for genome-wide quantification of splicing efficiency. BMC Bioinformatics 22:368, 2021.
Contact
For contacting me, please write to:
-
Prof. Rosario M. Piro
Politecnico di Milano
Dipartimento di Elettronica, Informazione e Bioingegneria (DEIB)
Via Ponzio 34/5
20133 Milano, Italy
Phone: +39 02 2399 3538
E-mail: rosariomichael.piro -AT- polimi.it
Institutional website: Piro @ DEIB
Impressum/Legal notice according to German legislation:
[This website is hosted in Germany...]
Responsible for the content of this website according to § 5 TMG (German Act for Telecommunications Media Services) and § 55 Abs. 2 RStV (German Interstate Broadcasting Agreement):
Prof. Rosario M. Piro
Politecnico di Milano
Dipartimento di Elettronica, Informazione e Bioingegneria (DEIB)
Via Ponzio 34/5
20133 Milano, Italy
E-mail: rosariomichael.piro -AT- polimi.it
Liability for the content of linked websites:
This website contains links to external websites on whose content I have no influence and for which I thus cannot take any responsibility. The responsibility for the content of linked websites lies with the providers or administrators of the respective websites. Linked websites have been checked for a possible infringement of laws upon creation of the link. No infringement could be identified when the links were created. A permanent control of the content of the linked websites is not reasonable without a tangible suspicion of the violation of a law. If I should be notified or otherwise become aware of violations of law by the content of a linked website, I will immediately remove the corresponding link.
 R.M.Piro
R.M.Piro