Computational Neuropathology & Oncology (CNO) Research Group
Politecnico di Milano
Dipartimento di Elettronica, Informazione e Bioingegneria (DEIB)
Via Ponzio 34/5, 20133 Milano, Italy
Research
Basic medical research frequently involves massive data production, such as the determination of genomic sequences and genome-wide gene expression levels by high throughput sequencing. Consequently, the importance of computational biology for medical research has constantly increased over the last decades. Both the analysis of medical data and the development of novel computational methods for this purpose are essential to keep pace with fast evolving experimental techniques and their application to growing numbers of patient samples.
Our Computational Neuropathology & Oncology (CNO) research group (established in March 2015 at Freie Universität Berlin and moved to Polictecnico di Milano in February 2020) develops novel computational approaches and applies existing methods with a primary focus on computational neuropathology, including cancer genetics/genomics and regulatory changes in metabolic and signaling networks of brain tumors, as well as topics related to the identification and the prioritization of candidate genes for human hereditary disorders of the central nervous system. Our research interest further extends from brain cancer to computational oncology in general, including other solid tumors and leukemias.
People
Principal Investigator:
 |
Rosario M. Piro, PhD
E-mail: rosariomichael.piro -AT- polimi.it |
PhD Students:
 |
|
Students:
 |
|
Alumni - Politecnico di Milano:
 |
Sabrina Anna Tidjani
Thesis: Mutational Effects on CTCF Binding Sites: Characterizing Alterations in Directionality |
 |
Aleyna Kazik
|
 |
Vittoria Esposito
Thesis: Relationship between the activity of tumor-specific mutational processes and the mutation status of cancer genes |
 |
Luca Procaccio
Thesis: A Wavelet-based approach for copy number variant calling |
Alumni - Freie Universität Berlin:
 |
Verônica Rodrigues de Melo Costa
Thesis: Genome-wide determination of splicing efficiency and dynamics from RNA-seq data |
 |
Rafael Alberdi Vallejo
Thesis: Vorhersagen von krankheitsassoziierten Genen mittels hochauflösenden Genexpressionsprofilen des Gehirns |
 |
Tobias Balzer
Thesis: Normalisation of mutational signatures for the application on specific genomic sites |
 |
Calvinna Caswara
Thesis: Evaluating voting-based consensus clustering of tumor samples using expression patterns from individual metabolic pathways |
 |
Willie Ekaputra
Thesis: Analysis of the differential co-expression of protein complexes in breast cancer |
 |
Franziska Fritz
Thesis: Relationship between recurrently mutated tumor suppressor genes and mutational processes |
 |
Sharen Nicole Heinig
Thesis: Mutability of oncogenes and tumor suppressor genes by different mutational processes |
 |
Jan Hoffschulte
Thesis: Evaluierung eines Peak Calling basierten Ansatzes zur Identifizierung intronischer Transkription in RNA-Seq Datensätzen |
 |
Marc Horlacher
Thesis: Analyse viraler Proteine im Kontext des humanen Protein-Interaktions-Netzwerks |
 |
Jacqueline Krohn
Thesis: Evaluierung der Krankheitsvorhersage mittels 3D-Genexpressionsdaten durch Unterscheidung einzelner Gehirnregionen |
 |
Sandra Krüger
Thesis (B.Sc.): Identifizierung von Mutationssignaturen in einzelnen Tumoren |
 |
Nina Kubiessa
Thesis: Verbesserung von Krankheitsgenvorhersagen basiert auf 3D-Genexpressionsprofilen |
 |
Nathaly Reimert
Thesis: Impact of mutational processes on the mutation frequency of tumor suppressors |
 |
Bonian Riebe
Thesis: Evaluierung einer Clustering-basierten Pathway-Analyse für mehrere Tumorklassen |
 |
Anja Seidel
Thesis: Evaluierung der Haar-Wavelet-Transformation für den Vergleich von 3D-Genexpressionsprofilen zur Vorhersage von krankheitsassoziierten Genen |
 |
Arsene Martinien Wabo Ngouambe
Thesis: Identifizierung von rekurrenten somatischen Mutationen in kurzen Introns |
Collaborations
(In alphabetical order)
-
Muhammad Sirajo Abdullahi
Department of Mathematics, Usmanu Danfodiyo University, Sokoto, Nigeria
-
Stefano Ceri
Dipartimento di Elettronica, Informazione e Bioingegneria (DEIB), Politecnico di Milano, Milan, Italy
-
Rainer König
Center for Sepsis Control & Care (CSCC), Jena University Hospital, Jena, Germany
-
Colin Logie
Department of Molecular Biology, Radboud University, Nijmegen, The Netherlands
-
Annalisa Marsico
Institute of Computational Biology (ICB), Helmholtz Zentrum München, Munich, Germany
-
Marco Masseroli
Dipartimento di Elettronica, Informazione e Bioingegneria (DEIB), Politecnico di Milano, Milan, Italy
-
Pietro Pinoli
Dipartimento di Elettronica, Informazione e Bioingegneria (DEIB), Politecnico di Milano, Milan, Italy
-
Kitiporn Plaimas
Department of Mathematics and Computer Science, Chulalongkorn University, Bangkok, Thailand
-
Apichat Suratanee
Department of Mathematics, King Mongkut's University of Technology North Bangkok, Thailand
Publications
(CNO group members are highlighted)
Highlights
2024
-
[Research article] Abdullahi MS, Suratanee A, Piro RM*, Plaimas K*. (*= senior authors)
Persistent Homology Identifies Pathways Associated with Hepatocellular Carcinoma from Peripheral Blood Samples.
Mathematics 12(5):725, 2024.
2023
-
[Research article] Reuss DE, Downing SM, Camacho CV, Wang Y-D, Piro RM, Herold-Mende C, Wang Z-Q, Hofmann TG, Sahm F, von Deimling A, McKinnon PJ, Frappart P-O.
Simultaneous Nbs1 and p53 inactivation in neural progenitors triggers High-Grade Gliomas (HGG).
Neuropathol Appl Neurobiol 49(4):e12915, 2023.
2021
-
[Research article] de Melo Costa VR, Pfeuffer J, Louloupi A, Ørom UAV, Piro RM.
SPLICE-q: a Python tool for genome-wide quantification of splicing efficiency.
BMC Bioinformatics 22:368, 2021.
-
[Research article] Epping L, Walther B, Piro RM, Knüver M-T, Huber C, Thürmer A, Flieger A, Fruth A, Janecko N, Wieler LH, Stingl K, Semmler T.
Genome-wide insights into population structure and host specificity of Campylobacter jejuni.
Scientific Reports 11(1):10358, 2021.
-
[Research article] Gulino A, Stamoulakatou E, Piro RM.
MutViz 2.0: visual analysis of somatic mutations and the impact of mutational signatures on selected genomic regions.
NAR Cancer 3(2):zcab012, 2021.
2020
-
[Book chapter / review article] Piro RM.
Sequencing technologies for epigenetics: from basics to applications.
Chapter 7 in: Kabelitz D, Bhat J (eds.) Epigenetics of the Immune System (Translational Epigenetics volume 16), pp. 161-183. Academic Press (Elsevier). ISBN: 978-0-1281-7964-2.
-
[Research article] Soni H, Bode J, Nguyen CDL, Puccio L, Neßling M, Piro RM, Bub J, Phillips E, Ahrends R, Eipper BA, Tews B, Goidts V.
PERK-mediated expression of peptidylglycine α-amidating monooxygenase supports angiogenesis in glioblastoma.
Oncogenesis 9(2):18, 2020.
-
[Research article] Stamoulakatou E, Pinoli P, Ceri S, Piro RM.
Impact of mutational signatures on microRNA and their response elements.
Pac Symp Biocomput 25:250-261, 2020.
2019
-
[Research article] Pinoli P, Stamoulakatou E, Ceri S, Piro RM.
Deleterious impact of mutational processes on transcription factor binding sites in human cancer.
Proceedings of the 19th IEEE International Conference on Bioinformatics and Bioengineering (BIBE), pp. 188-193, Athens, Greece, October 28-30, 2019.
-
[Research article] Krüger S, Piro RM.
decompTumor2Sig: identification of mutational signatures active in individual tumors.
BMC Bioinformatics 20(Suppl 4):152, 2019.
-
[Book chapter / review article] Piro RM, Marsico A.
Network-based methods and other approaches for predicting lncRNA functions and disease associations.
Methods in Molecular Biology 1912:301-321, 2019.
Chapter 12 in: Lai X, Gupta SK, Vera J (eds.), Computational Biology of Non-Coding RNA (MIMB, volume 1912), pp. 301-321. Humana Press/Springer, New York, NY, USA. ISBN: 978-1-4939-8981-2.
2018
-
[Research article] Gröbner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA, Johann PD, Balasubramanian GP, Segura-Wang M, Brabetz S, Bender S, Hutter B, Sturm D, Pfaff E, Hübschmann D, Zipprich G, Heinold M, Eils J, Lawerenz C, Erkek S, Lambo S, Waszak S, Blattmann C, Borkhardt A, Kuhlen M, Eggert A, Fulda S, Gessler M, Wegert J, Kappler R, Baumhoer D, Burdach S, Kirschner-Schwabe R, Kontny U, Kulozik AE, Lohmann D, Hettmer S, Eckert C, Bielack S, Nathrath M, Niemeyer C, Richter GH, Schulte J, Siebert R, Westermann F, Molenaar JJ, Vassal G, Witt H, Burkhardt B, Kratz CP, Witt O, van Tilburg CM, Kramm CM, Fleischhack G, Dirksen U, Rutkowski S, Frühwald M, von Hoff K, Wolf S, Klingebiel T, Koscielniak E, Landgraf P, Koster J, Resnick AC, Zhang J, Liu Y, Zhou X, Waanders AJ, Zwijnenburg DA, Raman P, Brors B, Weber UD, Northcott PA, Pajtler KW, Kool M, Piro RM, Korbel JO, Schlesner M, Eils R, Jones DTW, Lichter P, Chavez L, Zapatka M, Pfister SM; ICGC PedBrain-Seq Project; ICGC MMML-Seq Project.
The landscape of genomic alterations across childhood cancers.
Nature 555(7696):321-327, 2018.
2017
-
[Extended abstract] Krüger S, Piro RM.
Identification of mutational signatures active in individual tumors.
Proceedings of the NETTAB 2017 Workshop on Methods, Tools & Platforms for Personalized Medicine in the Big Data Era, Palermo, Italy, October 16-18, 2017.
PeerJ Preprints 5:e3257v1, 2017.
-
[Book] Robinson PN, Piro RM, Jäger M.
Computational Exome and Genome Analysis.
Chapman & Hall/CRC Mathematical and Computational Biology series. CRC Press (Taylor & Francis Group), Boca Raton, FL, USA. ISBN: 978-1-4987-7598-4.
-
[Research article] Schweizer Y, Meszaros Z, Jones DTW, Koelsche C, Boudalil M, Fiesel P, Schrimpf D, Piro RM, Brehmer S, von Deimling A, Kerl U, Seiz-Rosenhagen M, Capper D.
Molecular transition of an adult low-grade brain tumor to an atypical teratoid/rhabdoid tumor over a time-course of 14 years.
Journal of Neuropathology & Experimental Neurology 76(8):655-664, 2017.
2016
-
[Research article] Hutter S, Piro RM, Waszak SM, Kehrer-Sawatzki H, Friedrich RE, Lassaletta A, Witt O, Korbel JO, Lichter P, Schuhmann MU, Pfister SM, Tabori U, Mautner VF, Jones DTW.
No correlation between NF1 mutation position and risk of optic pathway glioma in 77 unrelated NF1 patients.
Human Genetics 135(5):469-475, 2016.
Downloads
decompTumor2Sig:
The decompTumor2Sig R package uses quadratic programming to decompose the mutation catalog from an individual tumor sample (or multiple individual tumor samples) into a set of given mutational signatures (either the Alexandrov model or the Shiraishi model), computing weights that reflect the contributions of the signatures to the mutation load of the tumor.
Citing decompTumor2Sig: Krüger S, Piro RM. decompTumor2Sig: identification of mutational signatures active in individual tumors. BMC Bioinformatics 20(Suppl 4):152, 2019.
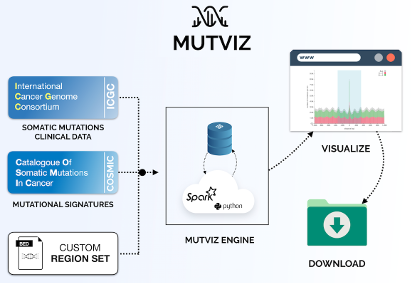
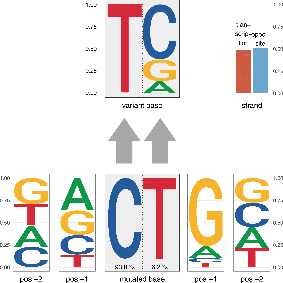
MutViz 2.0:
MutViz is a web tool which helps to identify enrichments of somatic mutations (single nucelotide variants) falling on predefined or user-specified sets of small genomic regions, such as transcription factor binding sites, promoters and others. MutViz is developed within the Data-driven Genomic Computing (GeCo) project.
Citing MutViz 2.0: Gulino A, Stamoulakatou E, Piro RM. MutViz 2.0: visual analysis of somatic mutations and the impact of mutational signatures on selected genomic regions. NAR Cancer 3(2):zcab012, 2021.
SPLICE-q:
SPLICE-q is a Python tool for genome-wide SPLICing Efficiency quantification from RNA-seq data. I takes a BAM file of mapped, strand-specific RNA-seq reads and a GTF gene annotation file and computes one of two available measurements of splicing efficiency for individual introns.
Citing SPLICE-q: de Melo Costa VR, Pfeuffer J, Louloupi A, Ørom UAV, Piro RM. SPLICE-q: a Python tool for genome-wide quantification of splicing efficiency. BMC Bioinformatics 22:368, 2021.
[For further downloads, see R.M.Piro → Downloads]
News
We offer thesis projects for M.Sc. students at Politecnico di Milano!
2021-10-12
CFP: please consider sumitting an article to special issue "Computational Analysis of Mutation Patterns and Mutational Signatures in Cancer" of the journal Biomolecules (Guest editor: R. M. Piro; deadline: 31 December 2021).
2021-04-09
|
New research article published in NAR Cancer: MutViz 2.0: visual analysis of somatic mutations and the impact of mutational signatures on selected genomic regions. |
2020-06-05
New review article/book chapter published in the Translational Epigenetics Series of Elsevier/Academic Press: "Sequencing technologies for epigenetics: from basics to applications".
2020-02-17
The group has officially moved from Freie Universität Berlin to the Department of Electronics, Information and Bioingeneering (DEIB) at Politecnico di Milano.
2019-05-03
|
Our R package decompTumor2Sig has just been released with Bioconductor 3.9! |
2020-01-03
New research article to be presented at the Pacific Symposium of Biocomputing 2020 in Hawaii, USA: "Impact of mutational signatures on microRNA and their response elements".
2019-10-28
New research article to be presented at the 19th IEEE International Conference on Bioinformatics and Bioengineering (BIBE) 2019 in Athens, Greece: "Deleterious impact of mutational processes on transcription factor binding sites in human cancer".
2019-04-18
New research article: "decompTumor2Sig: identification of mutational signatures active in individual tumors".
2019-01-12
New review article/book chapter: "Network-based methods and other approaches for predicting lncRNA functions and disease associations".
2017-08-10
|
The CRC Press book Computational Exome and Genome Analysis is finally being printed! |
2017-07-13
Our talk submitted to the NETTAB 2017 workshop has been accepted!
2017-01-12
Inivited talk at the IEEE conference Confluence-2017, Noida, India.
2015-03-16
CNO research group established at Freie Universität Berlin and Charité-Universitätsmedizin Berlin, Germany.
Contact
For contacting the research group, please write to:
-
Prof. Rosario M. Piro
Politecnico di Milano
Dipartimento di Elettronica, Informazione e Bioingegneria (DEIB)
Building 20
Via Ponzio 34/5
20133 Milano, Italy
Phone: +39 02 2399 3538
E-mail: rosariomichael.piro -AT- polimi.it
Institutional website: Piro @ DEIB
Where to find us:
(Note: Building 20 is the next building east of the "Mario Silvestri" building shown on the embedded map.)
Impressum/Legal notice according to German legislation:
[This website is hosted in Germany...]
Responsible for the content of this website according to § 5 TMG (German Act for Telecommunications Media Services) and § 55 Abs. 2 RStV (German Interstate Broadcasting Agreement):
Prof. Rosario M. Piro
Politecnico di Milano
Dipartimento di Elettronica, Informazione e Bioingegneria (DEIB)
Via Ponzio 34/5
20133 Milano, Italy
E-mail: rosariomichael.piro -AT- polimi.it
Liability for the content of linked websites:
This website contains links to external websites on whose content I have no influence and for which I thus cannot take any responsibility. The responsibility for the content of linked websites lies with the providers or administrators of the respective websites. Linked websites have been checked for a possible infringement of laws upon creation of the link. No infringement could be identified when the links were created. A permanent control of the content of the linked websites is not reasonable without a tangible suspicion of the violation of a law. If I should be notified or otherwise become aware of violations of law by the content of a linked website, I will immediately remove the corresponding link.
 R.M.Piro
R.M.Piro