About PathWave
PathWave enables the identification of disease specific regulation patterns by combining gene expression data and network topology. PathWave is preferable to classical enrichment tests as it offers a much higher sensitivity for detecting such functionally related regulation patterns. Additionally, it is more precise to a permutation based enrichment method (Schramm et al 2010).
Citing PathWave:
Please cite Piro et al (2014) for PathWave version 2.1, Schramm et al (2010) for PathWave version 1.0, and both papers (Schramm et al 2010; Piro et al 2014) for PathWave in general (see Publications).
Download
PathWave version 2.1 (Piro et al. 2014):
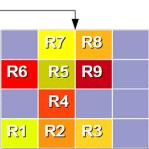
- GridArranger v1.0 - used to arrange pathways into compact 2D lattice grids (described in Schramm et al 2010; Oswald et al., 2012; Piro et al 2014; see Publications).
- GridArranger v1.0 README - basic information on installation and usage.
- ABACUS v2.4-alpha - required by GridArranger; this package is provided with kind permission of the original authors at the University of Cologne, Germany, because newer versions of ABACUS (available at the ABACUS webiste) are not compatible with GridArranger.
The R package does already include all necessary data for BiGG/Human recon 1 (H. sapiens) and KEGG (H. sapiens, M. musculus, D. melanogaster, C. elegans, D. rerio, E. coli).
Additional software for PathWave version 2.1:
The following software is needed only to preprocess further pathway data sets from KEGG (KGML) or BiGG (SBML), not for applying PathWave to already available, preprocessed pathway data.
GridArranger:
Note: GridArranger is also included as external code in the PathWave 2.1 R package. We provide it here as an independent package because it may be useful for other purposes.
ABACUS:
PathWave version 1.0 (Schramm et al. 2010):
[PathWave 1.0 was a collection of scripts and data files, rather than a package.]
Publications
Please cite Piro et al (2014) for PathWave version 2.1, Schramm et al (2010) for PathWave version 1.0, and both papers (Schramm et al 2010; Piro et al 2014) for PathWave in general.
Publications describing PathWave:
Network topology-based detection of differential gene regulation and regulatory switches in cell metabolism and signaling
BMC Systems Biology 8:56, 2014.
Rosario M. Piro (1,2,#), Stefan Wiesberg (3), Gunnar Schramm (1,2), Nico Rebel (3), Marcus Oswald (1,4,5), Roland Eils (1,2), Gerhard Reinelt (3), and Rainer König (1,2,4,5*)
(1) Division of Theoretical Bioinformatics, German Cancer Research Center (Deutsches Krebsforschungszentrum, DKFZ), Heidelberg, Germany
(2) Department of Bioinformatics and Functional Genomics, Institute of Pharmacy and Molecular Biotechnology, BioQuant, University of Heidelberg, Germany
(3) Institute of Computer Science and Interdisciplinary Center for Scientific Computing, University of Heidelberg, Germany
(4) Center for Sepsis Control and Care, University Hospital Jena, Jena, Germany
(5) Hans-Knöll-Institute (HKI), Jena, Germany
(#) Current address: Division of Molecular Genetics, German Cancer Research Center (Deutsches Krebsforschungszentrum, DKFZ), Heidelberg, Germany
(*) corresponding author: rainer.koenig@uni-jena.de
PathWave: discovering patterns of differentially regulated enzymes in metabolic pathways
Bioinformatics 26(9):1225-1231, 2010.
Gunnar Schramm (1,2), Stefan Wiesberg (3,4), Nicolle Diessl (1), Anna-Lena Kranz (1,2), Vitalia Sagulenko (5), Marcus Oswald (3,4), Gerhard Reinelt (3), Frank Westermann (5), Roland Eils (1,2,*) and Rainer König (1,2,*)
(1) Department of Bioinformatics and Functional Genomics, Institute of Pharmacy and Molecular Biotechnology, and Bioquant, Im Neuenheimer Feld 267, 69120 Heidelberg, Germany
(2) Department of Theoretical Bioinformatics, German Cancer Research Center (DKFZ), Im Neuenheimer Feld 280, 69120 Heidelberg, Germany
(3) Institute of Computer Science, University of Heidelberg, 69120 Heidelberg, Germany
(4) Interdisciplinary Center for Scientific Computing, University of Heidelberg, 69120 Heidelberg, Germany
(5) Department of Tumor Genetics, German Cancer Research Center (DKFZ), Im Neuenheimer Feld 280, 69120 Heidelberg, Germany
(*) corresponding authors: r.eils@dkfz.de and rainer.koenig@uni-jena.de
Mathematical foundation for grid optimization in PathWave:
Exact solution of the 2-dimensional grid arrangement problem
Discrete Optimization 9:189-199, 2012.
Marcus Oswald (1,2), Gerhard Reinelt (1), Stefan Wiesberg (1,2)
(1) Institute of Computer Science, University of Heidelberg, 69120 Heidelberg, Germany
(2) Interdisciplinary Center for Scientific Computing, University of Heidelberg, 69120 Heidelberg, Germany
Contact
For questions about PathWave, please write to:
-
Prof. Rosario M. Piro
Politecnico di Milano
Dipartimento di Elettronica, Informazione e Bioingegneria (DEIB)
Via Ponzio 34/5
20133 Milano, Italy
Phone: +39 02 2399 3538
E-mail: rosariomichael.piro -AT- polimi.it
Institutional website: Piro @ DEIB
Impressum/Legal notice according to German legislation:
[This website is hosted in Germany...]
Responsible for the content of this website according to § 5 TMG (German Act for Telecommunications Media Services) and § 55 Abs. 2 RStV (German Interstate Broadcasting Agreement):
Prof. Rosario M. Piro
Politecnico di Milano
Dipartimento di Elettronica, Informazione e Bioingegneria (DEIB)
Via Ponzio 34/5
20133 Milano, Italy
E-mail: rosariomichael.piro -AT- polimi.it
Liability for the content of linked websites:
This website contains links to external websites on whose content I have no influence and for which I thus cannot take any responsibility. The responsibility for the content of linked websites lies with the providers or administrators of the respective websites. Linked websites have been checked for a possible infringement of laws upon creation of the link. No infringement could be identified when the links were created. A permanent control of the content of the linked websites is not reasonable without a tangible suspicion of the violation of a law. If I should be notified or otherwise become aware of violations of law by the content of a linked website, I will immediately remove the corresponding link.
 R.M.Piro
R.M.Piro